We are particularly interested in the Ten Eleven Translocation (TET) family of proteins that consists of 3 members; TET1, TET2, and TET3. TET proteins via their catalytic domain can oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and further to other oxi-mCs. Thus, they are instrumental in the process of DNA demethylation. Moreover, they can form complexes-via interaction- with other proteins. As a result, their impact on the regulation of gene expression is multifaceted; they can act as suppressors or activators in a context-dependent manner.
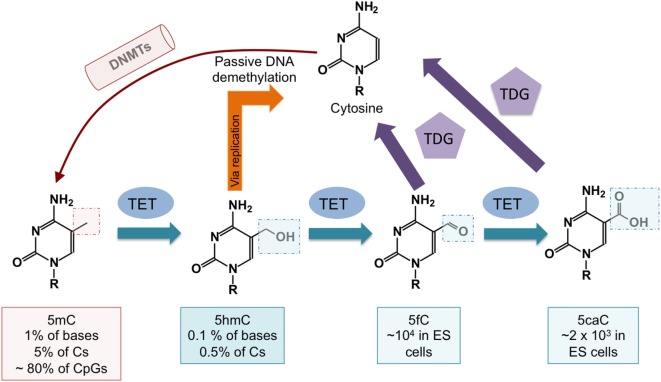
DNA demethylation pathways controlled by ten-eleven translocation (TET) proteins. DNA cytosine methylation is established by DNA methyltransferases (DNMTs). TET proteins oxidize 5mC to 5-hydroxymethylcytosine (5hmC). 5hmC can be passively diluted as a function of cell division. DNA demethylation can also occur via further oxidization of 5hmC by TET proteins to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Both 5fC and 5caC can be excised by thymine DNA glycosylase (TDG), resulting in their replacement with unmodified cytosine through base excision repair. (Tsagaratou et al., TET Methylcytosine Oxidases in T Cell and B Cell Development and Function. Front Immunol. 2017;8:220)
Our research so far has led us to a model of how the loss of TET2 and TET3 affects immune cell biology. In a wild-type precursor cell, TET2 and TET3 facilitate DNA demethylation and support the execution of a cell-specific gene expression program resulting in correct lineage specification and controlled cell proliferation. In Tet2/3 DKO precursor cells, the gene expression program is profoundly altered with consequent lineage skewing, aberrant proliferation, and malignant transformation. Next, we aim to decipher how each TET protein exerts its role in immune regulation and malignant transformation.

(Tsagaratou et al., TET Methylcytosine Oxidases in T Cell and B Cell Development and Function. Front Immunol. 2017;8:220).


